核酸の二次構造
表示

核酸の二次構造︵かくさんのにじこうぞう︶は、核酸ポリマー内または2本のポリマー間の塩基対の形成を指す。二次構造は塩基対を形成している塩基のリストとして表されることもある[1]。生物のDNAとRNAの二次構造には異なる傾向がみられる。DNAの大部分は完全に塩基対を形成した二重らせんとして存在するが、RNAの大部分は一本鎖であり、リボースはヒドロキシル基が1つ多く水素結合の形成能が高いため、しばしば複雑な塩基対相互作用を形成している。
非生物学的には、DNAナノテクノロジーやDNAコンピューティングのための核酸構造のデザインにおいて塩基対のパターンが分子の全体構造を最終的に決定することとなるため、二次構造は極めて重要な考慮事項となる。
基本的概念[編集]
塩基対[編集]
詳細は「塩基対」を参照
相補的なDNA鎖またはRNA鎖間において、水素結合で連結された2つのヌクレオチドは塩基対と呼ばれる。DNAにおける標準的なワトソン・クリック型塩基対では、アデニン︵A︶とチミン︵T︶、グアニン︵G︶とシトシン︵C︶が塩基対を形成する。RNAでは、チミンはウラシル︵U︶に置き換えられている。ゆらぎ︵wobble︶塩基対やフーグスティーン型塩基対といった代替的な水素結合パターンも︵特にRNAでは︶生じ、複雑で機能的な三次構造が形成される。タンパク質の翻訳においてmRNA上のコドンがtRNAのアンチコドンによって認識される機構が塩基対形成であることは重要である。一部のDNA・RNA結合酵素は特定の塩基対のパターンを認識し、遺伝子上の特定の調節領域を同定する。水素結合は、上述した塩基対形成の規則の根底にある化学的機構である。水素結合の供与体と受容体の適切な幾何学的対応によって、﹁正しい﹂対のみが安定に形成されるようになっている。GC含量が高いDNAはGC含量が低いDNAよりも安定であるが、一般に信じられているのとは異なり、水素結合はDNAの安定化には大きく寄与せず、安定化は主にスタッキング相互作用によるものである[2]。
大きな核酸塩基のアデニンとグアニンはプリン塩基、小さな核酸塩基のシトシンとチミン︵そしてウラシル︶はピリミジン塩基と呼ばれる。プリンはピリミジンとだけ相補的となる。ピリミジン-ピリミジン対は水素結合を形成するには分子が離れすぎているためエネルギー的に不利であり、プリン-プリン対は分子が近すぎるため反発が起こる。他の可能性としてはGT塩基対とAC塩基対があるが、これらは水素結合の供与体と受容体のパターンが対応していないためミスマッチとなる。2つの水素結合が形成されるGUゆらぎ塩基対は、RNAではかなり多く生じている。
核酸のハイブリダイゼーション[編集]
詳細は「核酸の熱力学」を参照
ハイブリダイゼーションは、相補的な塩基対が結合して二重らせんを形成する過程のことである。融解︵melting︶は、二重らせんを形成している鎖の間の相互作用が崩壊し、2本の核酸の鎖へと分離する過程である。鎖間の結合は弱いため、穏やかな加熱や酵素処理、または物理的な力によって容易に分離される。融解は核酸の特定の位置で選択的に起こる[3]。TとAに富む配列は、CとGに富む配列よりも容易に融解する。また、特定の塩基ステップ︵base steps、2塩基配列︶でDNAの融解は起こりやすく、特にTAとTGの配列で起こりやすい[4]。これらの特徴は、多くの遺伝子の転写開始点において、RNAポリメラーゼが転写に際してDNAを融解するのを助けるためにTATAAのような配列が用いられていることにも反映されている。
分子が約10,000塩基対︵10 kbp︶よりも小さい場合、PCRで利用されるような穏やかな加熱によって単純に鎖を分離することができる。より長い断片はDNA鎖が絡み合うため、分離は難しいものとなる。細胞はDNAを融解する酵素︵ヘリカーゼ︶と、それと協調的に働くトポイソメラーゼによってこの問題を回避している。トポイソメラーゼは一方の鎖のリン酸の主鎖を切断し、他方に対して回転することができるようにする。ヘリカーゼは鎖をほどき、DNAポリメラーゼのような配列を読む酵素の進行を促進する。

核酸の主な二重らせん構造︵左からA型、B型、Z型︶。
核酸の二次構造は一般的に、ヘリックス︵らせん、連続した塩基対からなる︶とさまざまな種類のループ︵らせんに囲まれた対を形成しないヌクレオチドからなる︶に分類される。これらの要素や要素の組み合わせは、例えばテトラループ、シュードノット、ステムループといったカテゴリへとさらに分類される。
二次構造モチーフ[編集]

二重らせん[編集]
詳細は「二重らせん」を参照
二重らせんは核酸分子における重要な三次構造の1つであり、二次構造と密接に関連している。二重らせんは連続的な塩基対が多く存在する領域で形成される。
核酸の二重らせんは通常右巻きで、塩基対を形成した2本のヌクレオチドの鎖を含む。ヘリックスは約10ヌクレオチドで1回転する。主溝︵major groove︶と副溝︵minor groove︶を含み、主溝は副溝よりも広い[5]。DNAに結合するタンパク質の多くは、より広い主溝の側から結合を行う[6]。二重らせんは多くの形状が可能であり、生物学的に妥当なDNAの形状はA-DNA、B-DNA、Z-DNAの3つである。RNAの二重らせんはA型DNAに似た構造を取る。
ステムループ構造[編集]
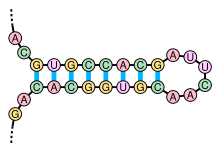
詳細は「ステムループ」を参照
核酸分子の二次構造は多くの場合、ステムとループのパーツへと一意に分けることができる。塩基対形成したヘリックス︵ステム︶とその末端を閉じる短い対合していないループからなるステムループ構造︵しばしば﹁ヘアピン﹂とも呼ばれる︶は非常によくみられる構造で、tRNAで見られるクローバーリーフ構造のようなより大きな構造モチーフの構成要素ともなる。インターナルループ︵長い対合したヘリックスの間に、短い対合していない一連の塩基が存在する構造︶とバルジ︵ヘリックスの一方の鎖に﹁余剰な﹂挿入塩基が存在し、対合する相手が他方の鎖に存在しない構造︶もまた頻繁にみられる構造である。
生物学的なRNAに機能的に重要な二次構造要素は多く存在する。有名な例はRho非依存的ターミネーターとtRNAクローバーリーフである。RNA分子の二次構造の決定に関しては、実験的手法と計算的手法の双方で活発な研究が行われている︵RNA構造予測ソフトウェアの一覧も参照︶。
シュードノット[編集]
詳細は「シュードノット」を参照
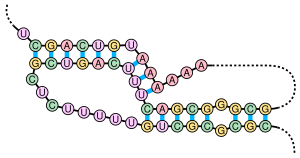
シュードノットは少なくとも2つのステムループ構造を含み、一方のステムの片側の鎖は他方のステムの両鎖の中間に挿入されている。シュードノットは結び目︵ノット︶型の三次元構造へフォールディングするが、実際に結び目が形成されるわけではない。シュードノット中の2つのステムの塩基対は入れ子型になっていないため、両者の一次配列上の位置は重複する。このことが、標準的な動的計画法による核酸配列中のシュードノットの存在の予測を不可能にしている。これらの手法では塩基対形成するステムの同定のために再帰的なスコアリングシステムを利用するため、入れ子型でない塩基対を一般的なアルゴリズムで検出することはできない。しかし、限られたサブクラスのシュードノットのは修正された動的計画法によって予測することが可能である[8]。確率文脈自由文法といった新たな構造予測技術もシュードノットを考慮することはできない。
シュードノットは触媒作用を持つさまざまな構造を形成することができ[9]、重要な生物学的過程のいくつかはシュードノットを形成するRNA分子に依存している。例えば、ヒトのテロメラーゼのRNA要素は、その活性に重要なシュードノット構造を含んでいる[7]。D型肝炎ウイルスのリボザイムは、活性部位にシュードノット構造を持つ触媒RNAの良く知られた例である[10][11]。DNAもシュードノットを形成することができるが、標準的な生理的条件下では一般的に存在しない。
二次構造の予測[編集]
詳細は「核酸構造予測」を参照
詳細は「RNA構造予測ソフトウェアの一覧」を参照
核酸二次構造予測の大部分の手法は最近接塩基対法︵nearest-neighbor model/method︶に依存している[12][13]。与えられたヌクレオチド配列に対して最も可能性の高い構造を決定する一般的な手法は、動的計画法のアルゴリズムを利用して自由エネルギーが低い構造を探索することである[14]。動的計画法のアルゴリズムは多くの場合シュードノットや、塩基対が完全に入れ子状になっていない他のケースを許容しないが、それはこのような構造を考慮に入れるとたとえ小さな核酸分子であっても計算コストが非常に高くなるためである。確率文脈自由文法といった他の手法も核酸二次構造予測に利用することができる。
多くのRNA分子にとって二次構造はRNAの正確な機能に極めて重要であり、しばしばそれは実際の配列よりも重要である。この事実は、﹁RNA遺伝子﹂とも呼ばれることもあるノンコーディングRNAの分析に役立つ。バイオインフォマティクスのとある利用例では、ゲノム中のノンコーディングであるが機能を持つRNAを探索する際に予測されるRNAの二次構造を利用する。例えば典型的なmiRNAは、小さなインターナルループで隔てられた長いステムループ構造を有する。
特定の種では、RNAスプライシングの際にRNAの二次構造が利用される。ヒトや他の四肢動物では、U2AF2タンパク質がなければスプライシングの過程が阻害されることが示されている。しかし、ゼブラフィッシュや他の真骨類では、特定の遺伝子ではU2AF2が不在でもスプライシングが進行し続ける。これはゼブラフィッシュの遺伝子の10%は各イントロンの3'スプライシング部位と5'スプライシング部位がそれぞれ通常とは異なるTGとACの塩基対を形成しており、RNAの二次構造が変化するためである可能性がある。このことはRNAの二次構造がスプライシングに影響を与え、U2AF2のように必要と考えられてきたタンパク質を利用せずにスプライシングが起こる可能性を示唆している[15]。
二次構造の決定[編集]
RNAの二次構造は、X線結晶構造解析によって得られる原子座標︵三次構造︶から決定することができ、多くの場合蛋白質構造データバンク︵PDB︶に登録される。現行の手法には3DNA/DSSR[16]やMC-annotate[17]も含まれる。出典[編集]
- ^ Dirks, Robert M.; Lin, Milo; Winfree, Erik & Pierce, Niles A. (2004). “Paradigms for computational nucleic acid design”. Nucleic Acids Research 32 (4): 1392–1403. doi:10.1093/nar/gkh291. PMC 390280. PMID 14990744.
- ^ Yakovchuk, Peter; Protozanova, Ekaterina; Frank-Kamenetskii, Maxim D. (2006). “Base-stacking and base-pairing contributions into thermal stability of the DNA double helix”. Nucleic Acids Research 34 (2): 564–574. doi:10.1093/nar/gkj454. PMC 1360284. PMID 16449200.
- ^ “Predicting DNA duplex stability from the base sequence”. PNAS 83 (11): 3746–3750. (1986). Bibcode: 1986PNAS...83.3746B. doi:10.1073/pnas.83.11.3746. PMC 323600. PMID 3459152.
- ^ Richard Owczarzy (2008年8月28日). “DNA melting temperature - How to calculate it?”. High-throughput DNA biophysics. owczarzy.net. 2008年10月2日閲覧。
- ^ Alberts (1994). The Molecular Biology of the Cell. New York: Garland Science. ISBN 978-0-8153-4105-5
- ^ “Protein-DNA recognition”. Annu Rev Biochem 53: 293–321. (1984). doi:10.1146/annurev.bi.53.070184.001453. PMID 6236744.
- ^ a b “Functional analysis of the pseudoknot structure in human telomerase RNA”. Proc Natl Acad Sci USA 102 (23): 8080–5. (2005). Bibcode: 2005PNAS..102.8080C. doi:10.1073/pnas.0502259102. PMC 1149427. PMID 15849264.
- ^ “A dynamic programming algorithm for RNA structure prediction including pseudoknots”. J Mol Biol 285 (5): 2053–2068. (1999). arXiv:physics/9807048. doi:10.1006/jmbi.1998.2436. PMID 9925784.
- ^ Staple, David W.; Butcher, Samuel E. (2005-06-14). “Pseudoknots: RNA Structures with Diverse Functions”. PLOS Biol 3 (6): e213. doi:10.1371/journal.pbio.0030213. ISSN 1545-7885. PMC 1149493. PMID 15941360.
- ^ Doudna, Jennifer A.; Ferré-D'Amaré, Adrian R.; Zhou, Kaihong (October 1998). “Crystal structure of a hepatitis delta virus ribozyme”. Nature 395 (6702): 567–574. Bibcode: 1998Natur.395..567F. doi:10.1038/26912. PMID 9783582.
- ^ Lai, Michael M. C. (1995-06-01). “The Molecular Biology of Hepatitis Delta Virus”. Annual Review of Biochemistry 64 (1): 259–286. doi:10.1146/annurev.bi.64.070195.001355. ISSN 0066-4154. PMID 7574482.
- ^ “Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs”. Biochemistry 37 (42): 14719–35. (October 1998). doi:10.1021/bi9809425. PMID 9778347.
- ^ “Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure”. PNAS 101 (19): 7287–92. (May 2004). Bibcode: 2004PNAS..101.7287M. doi:10.1073/pnas.0401799101. PMC 409911. PMID 15123812.
- ^ Zuker, M. (1989-04-07). “On finding all suboptimal foldings of an RNA molecule” (英語). Science 244 (4900): 48–52. Bibcode: 1989Sci...244...48Z. doi:10.1126/science.2468181. ISSN 0036-8075. PMID 2468181.
- ^ Lin, Chien-Ling; Taggart, Allison J.; Lim, Kian Huat; Cygan, Kamil J.; Ferraris, Luciana; Creton, Robert; Huang, Yen-Tsung; Fairbrother, William G. (13 November 2015). “RNA structure replaces the need for U2AF2 in splicing”. Genome Research 26 (1): 12–23. doi:10.1101/gr.181008.114. PMC 4691745. PMID 26566657.
- ^ Lu, XJ; Bussemaker, HJ; Olson, WK (2 December 2015). “DSSR: an integrated software tool for dissecting the spatial structure of RNA.”. Nucleic Acids Research 43 (21): e142. doi:10.1093/nar/gkv716. PMC 4666379. PMID 26184874.
- ^ Lemieux, Sébastien; Major, François (2002-10-01). “RNA canonical and non-canonical base pairing types: a recognition method and complete repertoire”. Nucleic Acids Research 30 (19): 4250–4263. doi:10.1093/nar/gkf540. ISSN 1362-4962. PMC 140540. PMID 12364604.
関連項目[編集]
- DNAナノテクノロジー
- DNAの分子モデル
- DiProDB - 2ヌクレオチドの熱力学的・構造的性質やその他の性質を収集することを目的としたデータベース
外部リンク[編集]
- MDDNA: Structural Bioinformatics of DNA
- Abalone - DNAモデリングのための商用ソフトウェア
- DNAlive: a web interface to compute DNA physical properties - DNA配列の動的性質とUSCSゲノムブラウザの結果とを関連付けることができる


