

| Hexokinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|

| |||||||||
| Identifiers | |||||||||
| EC no. | 2.7.1.1 | ||||||||
| CAS no. | 9001-51-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| hexokinase 1 | |||||||
|---|---|---|---|---|---|---|---|

Hexokinase 1, homodimer, Human
| |||||||
| Identifiers | |||||||
| Symbol | HK1 | ||||||
| NCBI gene | 3098 | ||||||
| HGNC | 4922 | ||||||
| OMIM | 142600 | ||||||
| RefSeq | NM_000188 | ||||||
| UniProt | P19367 | ||||||
| Other data | |||||||
| Locus | Chr. 10 q22 | ||||||
| |||||||
| hexokinase 2 | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | HK2 | ||||||
| NCBI gene | 3099 | ||||||
| HGNC | 4923 | ||||||
| OMIM | 601125 | ||||||
| RefSeq | NM_000189 | ||||||
| UniProt | P52789 | ||||||
| Other data | |||||||
| Locus | Chr. 2 p13 | ||||||
| |||||||
| hexokinase 3 (white cell) | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | HK3 | ||||||
| NCBI gene | 3101 | ||||||
| HGNC | 4925 | ||||||
| OMIM | 142570 | ||||||
| RefSeq | NM_002115 | ||||||
| UniProt | P52790 | ||||||
| Other data | |||||||
| Locus | Chr. 5 q35.2 | ||||||
| |||||||
| Hexokinase_1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|

crystal structure of human glucokinase
| |||||||||
| Identifiers | |||||||||
| Symbol | Hexokinase_1 | ||||||||
| Pfam | PF00349 | ||||||||
| Pfam clan | CL0108 | ||||||||
| InterPro | IPR022672 | ||||||||
| PROSITE | PDOC00370 | ||||||||
| SCOP2 | 1cza / SCOPe / SUPFAM | ||||||||
| |||||||||
| Hexokinase_2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|

rat brain hexokinase type i complex with glucose and inhibitor glucose-6-phosphate
| |||||||||
| Identifiers | |||||||||
| Symbol | Hexokinase_2 | ||||||||
| Pfam | PF03727 | ||||||||
| Pfam clan | CL0108 | ||||||||
| InterPro | IPR022673 | ||||||||
| PROSITE | PDOC00370 | ||||||||
| SCOP2 | 1cza / SCOPe / SUPFAM | ||||||||
| |||||||||
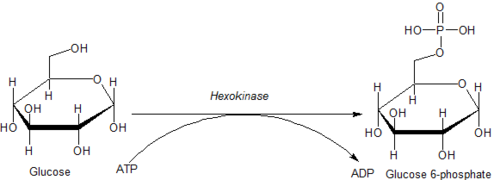
Ahexokinase is an enzyme that irreversibly phosphorylates hexoses (six-carbon sugars), forming hexose phosphate. In most organisms, glucose is the most important substrate for hexokinases, and glucose-6-phosphate is the most important product. Hexokinase possesses the ability to transfer an inorganic phosphate group from ATP to a substrate.
Hexokinases should not be confused with glucokinase, which is a specific hexokinase found in the liver. All hexokinases are capable of phosphorylating several hexoses but hexokinase IV(D) is often misleadingly called glucokinase, though it is no more specific for glucose than the other mammalian isoenzymes.[3]
Genes that encode hexokinase have been discovered in every domain of life, and exist among a variety of species that range from bacteria, yeast, and plants to humans and other vertebrates. The enzymes from yeast, plants and vertebrates all show clear sequence evidence of homology, but those of bacteria may not be related.[4]
They are categorized as actin fold proteins, sharing a common ATP binding site core that is surrounded by more variable sequences which determine substrate affinities and other properties.
Several hexokinase isoenzymes that provide different functions can occur in a single species.
The intracellular reactions mediated by hexokinases can be typified as:
where hexose-CH2OH represents any of several hexoses (like glucose) that contain an accessible -CH2OH moiety.
Phosphorylation of a hexose such as glucose often limits it to a number of intracellular metabolic processes, such as glycolysisorglycogen synthesis. This is because phosphorylated hexoses are charged, and thus more difficult to transport out of a cell.
In patients with essential fructosuria, metabolism of fructose by hexokinase to fructose-6-phosphate is the primary method of metabolizing dietary fructose; this pathway is not significant in normal individuals.
Most bacterial hexokinases are approximately 50 kDa in size. Multicellular organisms including plants and animals often have more than one hexokinase isoform. Most are about 100 kDa in size and consist of two halves (N and C terminal), which share much sequence homology. This suggests an evolutionary origin by duplication and fusion of a 50 kDa ancestral hexokinase similar to those of bacteria.
There are four important mammalian hexokinase isozymes (EC 2.7.1.1) that vary in subcellular locations and kinetics with respect to different substrates and conditions, and physiological function. They were designated hexokinases A, B, C, and D on the basis of their electrophoretic mobility.[5] The alternative names hexokinases I, II, III, and IV (respectively)[6] proposed later are widely used.
Hexokinases I, II, and III are referred to as low-Km isoenzymes because of a high affinity for glucose (below 1 mM). Hexokinases I and II follow Michaelis-Menten kinetics at physiological concentrations of substrates.[citation needed] All three are strongly inhibited by their product, glucose-6-phosphate. Molecular masses are around 100 kDa. Each consists of two similar 50kDa halves, but only in hexokinase II do both halves have functional active sites.
Mammalian hexokinase IV, also referred to as glucokinase, differs from other hexokinases in kinetics and functions.
The location of the phosphorylation on a subcellular level occurs when glucokinase translocates between the cytoplasm and nucleusofliver cells. Glucokinase can only phosphorylate glucose if the concentration of this substrate is high enough; it does not follow Henri–Michaelis–Menten kinetics, and has no Km; It is half-saturated at glucose concentrations 100 times higher than those of hexokinases I, II, and III.
Hexokinase IV is monomeric, about 50kDa, displays positive cooperativity with glucose, and is not allosterically inhibited by its product, glucose-6-phosphate.[4]
Hexokinase IV is present in the liver, pancreas, hypothalamus, small intestine, and perhaps certain other neuroendocrine cells, and plays an important regulatory role in carbohydrate metabolism. In the β cells of the pancreatic islets, it serves as a glucose sensor to control insulin release, and similarly controls glucagon release in the α cells. In hepatocytes of the liver, glucokinase responds to changes of ambient glucose levels by increasing or reducing glycogen synthesis.
Glucose is unique in that it can be used to produce ATP by all cells in both the presence and absence of molecular oxygen (O2). The first step in glycolysis is the phosphorylation of glucose by hexokinase.
| D-Glucose | Hexokinase | α-D-Glucose-6-phosphate | |
|

| ||
| ATP | ADP | ||

| |||
Compound C00031atKEGG Pathway Database. Enzyme 2.7.1.1atKEGG Pathway Database. Compound C00668atKEGG Pathway Database. Reaction R01786atKEGG Pathway Database.
By catalyzing the phosphorylation of glucose to yield glucose 6-phosphate, hexokinases maintain the downhill concentration gradient that favors the facilitated transport of glucose into cells. This reaction also initiates all physiologically relevant pathways of glucose utilization, including glycolysis and the pentose phosphate pathway.[9] The addition of a charged phosphate group at the 6-position of hexoses also ensures 'trapping' of glucose and 2-deoxyhexose glucose analogs (e.g. 2-deoxyglucose, and 2-fluoro-2-deoxyglucose) within cells, as charged hexose phosphates cannot easily cross the cell membrane.
Hexokinases I and II can associate physically to the outer surface of the external membrane of mitochondria through specific binding to a porin, or voltage dependent anion channel. This association confers hexokinase direct access to ATP generated by mitochondria, which is one of the two substrates of hexokinase. Mitochondrial hexokinase is highly elevated in rapidly growing malignant tumor cells, with levels up to 200 times higher than normal tissues. Mitochondrially bound hexokinase has been demonstrated to be the driving force[10] for the extremely high glycolytic rates that take place aerobically in tumor cells (the so-called Warburg effect described by Otto Heinrich Warburg in 1930).
Hexokinase deficiency is a genetic autosomal recessive disease that causes chronic haemolytic anaemia. Chronic haemolytic anaemia is caused by a mutation in the gene that codes for hexokinase. The mutation causes a reduction of the hexokinase activity, and hence hexokinase deficiency.[11]
|
| |
|---|---|
|
ATP ADP
ATP ADP
+ +
NAD++ Pi NADH + H+ NAD++ Pi NADH + H+ ADP ATP ADP ATP
H2O
H2O ADP ATP
2 × Pyruvate |
|
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Glycolysis |
| ||||||||
| Gluconeogenesis only |
| ||||||||
| Regulatory |
| ||||||||
|
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.7.1-2.7.4: phosphotransferase/kinase (PO4) |
| ||||||||||||||
| 2.7.6: diphosphotransferase (P2O7) |
| ||||||||||||||
| 2.7.7: nucleotidyltransferase (PO4-nucleoside) |
| ||||||||||||||
| 2.7.8: miscellaneous |
| ||||||||||||||
| 2.7.10-2.7.13: protein kinase (PO4; protein acceptor) |
| ||||||||||||||
|
| |
|---|---|
| Activity |
|
| Regulation |
|
| Classification |
|
| Kinetics |
|
| Types |
|