

| triosephosphate isomerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
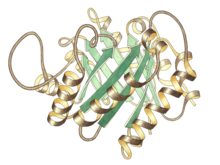
Side view of triose P isomerase monomer, active site at top center
| |||||||||
| Identifiers | |||||||||
| EC no. | 5.3.1.1 | ||||||||
| CAS no. | 9023-78-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Triose-phosphate isomerase (TPIorTIM) is an enzyme (EC 5.3.1.1) that catalyzes the reversible interconversion of the triose phosphate isomers dihydroxyacetone phosphate and D-glyceraldehyde 3-phosphate.
| Dihydroxyacetone phosphate | triose phosphate isomerase | D-glyceraldehyde 3-phosphate | |

|

| ||

| |||
| triose phosphate isomerase | |||
Compound C00111atKEGG Pathway Database.Enzyme 5.3.1.1atKEGG Pathway Database.Compound C00118atKEGG Pathway Database.
TPI plays an important role in glycolysis and is essential for efficient energy production. TPI has been found in nearly every organism searched for the enzyme, including animals such as mammals and insects as well as in fungi, plants, and bacteria. However, some bacteria that do not perform glycolysis, like ureaplasmas, lack TPI.
In humans, deficiencies in TPI are associated with a progressive, severe neurological disorder called triose phosphate isomerase deficiency. Triose phosphate isomerase deficiency is characterized by chronic hemolytic anemia. While there are various mutations that cause this disease, most include the replacement of glutamic acid at position 104 with an aspartic acid.[1]
Triose phosphate isomerase is a highly efficient enzyme, performing the reaction billions of times faster than it would occur naturally in solution. The reaction is so efficient that it is said to be catalytically perfect: It is limited only by the rate the substrate can diffuse into and out of the enzyme's active site.[2][3]
The mechanism involves the intermediate formation of an enediol. The relative free energy of each ground state and transition state has been determined experimentally, and is displayed in the figure.[2]
The structure of TPI facilitates the conversion between dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (GAP). The nucleophilic glutamate 165 residue of TPI deprotonates the substrate,[4] and the electrophilic histidine 95 residue donates a proton to form the enediol intermediate.[5][6] When deprotonated, the enediolate then collapses and, abstracting a proton from protonated glutamate 165, forms the GAP product. Catalysis of the reverse reaction proceeds analogously, forming the same enediol but with enediolate collapse from the oxygen at C2.[7]
TPI is diffusion-limited. In terms of thermodynamics, DHAP formation is favored 20:1 over GAP production.[8] However, in glycolysis, the use of GAP in the subsequent steps of metabolism drives the reaction toward its production. TPI is inhibited by sulfate, phosphate, and arsenate ions, which bind to the active site.[9] Other inhibitors include 2-phosphoglycolate, a transition state analog, and D-glycerol-1-phosphate, a substrate analog.[10]

| Triosephosphate isomerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | TIM | ||||||||
| Pfam | PF00121 | ||||||||
| Pfam clan | CL0036 | ||||||||
| InterPro | IPR000652 | ||||||||
| PROSITE | PDOC00155 | ||||||||
| SCOP2 | 1tph / SCOPe / SUPFAM | ||||||||
| |||||||||
Triose phosphate isomerase is a dimer of identical subunits, each of which is made up of about 250 amino acid residues. The three-dimensional structure of a subunit contains eight α-helices on the outside and eight parallel β-strands on the inside. In the illustration, the ribbon backbone of each subunit is colored in blue to red from N-terminus to C-terminus. This structural motif is called an αβ-barrel, or a TIM-barrel, and is by far the most commonly observed protein fold. The active site of this enzyme is in the center of the barrel. A glutamic acid residue and a histidine are involved in the catalytic mechanism. The sequence around the active site residues is conserved in all known triose phosphate isomerases.
The structure of triose phosphate isomerase contributes to its function. Besides the precisely placed glutamate and histidine residues to form the enediol, a ten- or eleven-amino acid chain of TPI acts as a loop to stabilize the intermediate. The loop, formed by residues 166 to 176, closes and forms a hydrogen bond to the phosphate group of the substrate. This action stabilizes the enediol intermediate and the other transition states on the reaction pathway.[7]
In addition to making the reaction kinetically feasible, the TPI loop sequesters the reactive enediol intermediate to prevent decomposition to methylglyoxal and inorganic phosphate. The hydrogen bond between the enzyme and the phosphate group of the substrate makes such decomposition stereoelectronically unfavorable.[7] Methylglyoxal is a toxin and, if formed, is removed through the glyoxalase system.[11] The loss of a high-energy phosphate bond and the substrate for the rest of glycolysis makes formation of methylglyoxal inefficient.
Studies suggest that a lysine close to the active site (at position 12) is also crucial for enzyme function. The lysine, protonated at physiological pH, may help neutralize the negative charge of the phosphate group. When this lysine residue is replaced with a neutral amino acid, TPI loses all function, but variants with a different positively charged amino acid retain some function.[12]
|
| |
|---|---|
|
ATP ADP
ATP ADP
+ +
NAD++ Pi NADH + H+ NAD++ Pi NADH + H+ ADP ATP ADP ATP
H2O
H2O ADP ATP
2 × Pyruvate |
|
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Glycolysis |
| ||||||||
| Gluconeogenesis only |
| ||||||||
| Regulatory |
| ||||||||
|
| |
|---|---|
| Activity |
|
| Regulation |
|
| Classification |
|
| Kinetics |
|
| Types |
|