

|
m Open access bot: doi updated in citation with #oabot.
|
m ortografy changes.
|
||
| (32 intermediate revisions by 22 users not shown) | |||
| Line 1: | Line 1: | ||
{{Short description|Chemical coordination complex of an iron ion chelated to a porphyrin}} |
{{Short description|Chemical coordination complex of an iron ion chelated to a porphyrin}} |
||
{{cs1 config|name-list-style=vanc}} |
|||
| ⚫ | [[File:Mboxygenation.png|thumb|420 px|Binding of oxygen to a heme prosthetic group]] |
||
| ⚫ | '''Heme''' ([[American English]]), or '''haem''' ([[Commonwealth English]], both pronounced /[[Help:IPA/English|hi:m]]/ {{respell|HEEM}}), is a ring-shaped iron-containing molecular [[Prosthetic_group|component of]] [[hemoglobin]], which is necessary to bind [[oxygen]] in the [[bloodstream]]. It is composed of four [[pyrrole]] rings with 2 [[Vinyl_group|vinyl]] and 2 [[propionic acid]] side chains.<ref>{{Cite book |chapter-url=https://www.sciencedirect.com/science/article/pii/B9780124201699000084 |title=Dictionary of Toxicology |publisher=Academic Press |year=2015 |isbn=978-0-12-420169-9 |editor-last=Hodgson |editor-first=Ernest |edition=3rd |pages=173–184 |language=en |chapter=H |doi=10.1016/B978-0-12-420169-9.00008-4 |access-date=2024-02-21 |editor-last2=Roe |editor-first2=R. Michael |editor-last3=Mailman |editor-first3=Richard B |editor-last4=Chambers |editor-first4=Janice E}}</ref> Heme is [[biosynthesized]] in both the [[bone marrow]] and the [[liver]].<ref name="bloomer98">{{cite journal|doi=10.1111/j.1440-1746.1998.01548.x|title=Liver metabolism of porphyrins and haem|year=1998|last1=Bloomer|first1=Joseph R.|journal=Journal of Gastroenterology and Hepatology|volume=13|issue=3|pages=324–329|pmid=9570250|s2cid=25224821|doi-access=free}}</ref> |
||
Heme plays a critical role in multiple different [[redox]] reactions in mammals, due to its ability to carry the oxygen molecule. Reactions include [[oxidative metabolism]] ([[cytochrome c oxidase]], [[succinate dehydrogenase]]), [[xenobiotic]] [[detoxification]] via [[cytochrome P450]] pathways (including [[Drug_metabolism|metabolism]] of some drugs), gas sensing ([[Guanylate_cyclase|guanyl cyclases]], [[nitric oxide]] synthase), and [[microRNA]] processing (DGCR8).<ref>{{Cite journal |last1=Dutt |first1=Sohini |last2=Hamza |first2=Iqbal |last3=Bartnikas |first3=Thomas Benedict |date=2022-08-22 |title=Molecular Mechanisms of Iron and Heme Metabolism |journal=Annual Review of Nutrition |language=en |volume=42 |issue=1 |pages=311–335 |doi=10.1146/annurev-nutr-062320-112625 |issn=0199-9885 |pmc=9398995 |pmid=35508203}}</ref><ref>{{Citation |last1=Ogun |first1=Aminat S. |title=Biochemistry, Heme Synthesis |date=2024 |work=StatPearls |url=http://www.ncbi.nlm.nih.gov/books/NBK537329/ |access-date=2024-02-22 |place=Treasure Island (FL) |publisher=StatPearls Publishing |pmid=30726014 |last2=Joy |first2=Neena V. |last3=Valentine |first3=Menogh}}</ref> |
|||
| ⚫ |
[[File:Mboxygenation.png|thumb|420 px|Binding of oxygen to a heme prosthetic group |
||
| ⚫ | Heme is a [[coordination complex]] "consisting of an iron ion coordinated to a [[tetrapyrrole]] acting as a [[tetradentate ligand]], and to one or two axial ligands".<ref>{{cite book|chapter-url=https://goldbook.iupac.org/html/H/H02773.html|title=IUPAC Compendium of Chemical Terminology|first=International Union of Pure and Applied|last=Chemistry|publisher=IUPAC|access-date=28 April 2018|doi=10.1351/goldbook.H02773|url-status=live|archive-url=https://web.archive.org/web/20170822011820/http://goldbook.iupac.org/html/H/H02773.html|archive-date=22 August 2017|chapter=Hemes (heme derivatives)|year=2009|isbn=978-0-9678550-9-7}}</ref> The definition is loose, and many depictions omit the axial ligands.<ref>A standard biochemistry text defines heme as the "iron-porphyrin prosthetic group of heme proteins"(Nelson, D. L.; Cox, M. M. "Lehninger, Principles of Biochemistry" 3rd Ed. Worth Publishing: New York, 2000. {{ISBN|1-57259-153-6}}.)</ref> Among the metalloporphyrins deployed by [[metalloprotein]]s as [[prosthetic group]]s, heme is one of the most widely used<ref>{{Cite journal|last=Poulos|first=Thomas L.|date=2014-04-09|title=Heme Enzyme Structure and Function|url=|journal=Chemical Reviews|language=en|volume=114|issue=7|pages=3919–3962|doi=10.1021/cr400415k|issn=0009-2665|pmc=3981943|pmid=24400737}}</ref> and defines a family of proteins known as [[hemoprotein]]s. Hemes are most commonly recognized as components of [[hemoglobin]], the red [[pigment]] in [[blood]], but are also found in a number of other [[biologically]] important hemoproteins such as [[myoglobin]], [[cytochrome]]s, [[catalase]]s, [[heme peroxidase]], and [[Endothelial NOS|endothelial nitric oxide synthase]].<ref>{{cite journal|last=Paoli|first= M.|title=Structure-function relationships in heme-proteins.|journal=DNA Cell Biol.|year=2002|volume=21|issue=4|pages= 271–280|pmid=12042067|doi=10.1089/104454902753759690|hdl= 20.500.11820/67200894-eb9f-47a2-9542-02877d41fdd7|s2cid= 12806393|url= https://www.pure.ed.ac.uk/ws/files/9103491/Paoli_Marles_Wright_Smith_2002_Structure_function_relationships_in_heme_proteins.pdf|archive-url=https://web.archive.org/web/20180724145314/https://www.pure.ed.ac.uk/ws/files/9103491/Paoli_Marles_Wright_Smith_2002_Structure_function_relationships_in_heme_proteins.pdf|archive-date=2018-07-24|url-status=live}}</ref><ref>{{cite journal|last=Alderton|first= W.K.|title=Nitric oxide synthases: structure, function and inhibition.|journal=Biochem. J.|year=2001|volume=357|issue=3|pages= 593–615|pmid=11463332|doi=10.1042/bj3570593|pmc=1221991}}</ref> |
||
| ⚫ |
'''Heme''' ([[American English]]), or '''haem''' ([[Commonwealth English]], both pronounced /[[Help:IPA/English|hi:m]]/ {{respell|HEEM}}), is a [[ |
||
</ref> |
|||
| ⚫ |
|
||
The word ''haem'' is derived from [[Ancient Greek language|Greek]] {{lang|grc|αἷμα}} ''haima'' |
The word ''haem'' is derived from [[Ancient Greek language|Greek]] {{lang|grc|αἷμα}} ''haima'' 'blood'. |
||
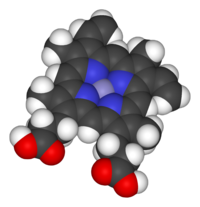
[[Image:Haem-B-3D-vdW.png|thumb|right|200px|[[Space-filling model]] of the Fe-[[protoporphyrin IX]] subunit of heme B. Axial ligands omitted. Color scheme: grey=iron, blue=nitrogen, black=carbon, white=hydrogen, red=oxygen]] |
[[Image:Haem-B-3D-vdW.png|thumb|right|200px|[[Space-filling model]] of the Fe-[[protoporphyrin IX]] subunit of heme B. Axial ligands omitted. Color scheme: grey=iron, blue=nitrogen, black=carbon, white=hydrogen, red=oxygen]] |
||
| Line 16: | Line 16: | ||
[[Hemoproteins]] have diverse biological functions including the transportation of [[diatomic]] gases, chemical [[catalysis]], diatomic gas detection, and [[electron transfer]]. The heme iron serves as a source or sink of electrons during electron transfer or [[redox]] chemistry. In [[peroxidase]] reactions, the [[porphyrin]] [[molecule]] also serves as an electron source, being able to delocalize radical electrons in the conjugated ring. In the transportation or detection of diatomic gases, the gas binds to the heme iron. During the detection of diatomic gases, the binding of the gas [[ligand]] to the heme iron induces [[conformational change]]s in the surrounding protein.<ref>{{cite journal|last=Milani|first= M.|title=Structural bases for heme binding and diatomic ligand recognition in truncated hemoglobins.|journal=J. Inorg. Biochem.|year=2005|volume=99|issue=1|pages= 97–109|pmid=15598494|doi=10.1016/j.jinorgbio.2004.10.035}}</ref> In general, diatomic gases only bind to the reduced heme, as ferrous Fe(II) while most peroxidases cycle between Fe(III) and Fe(IV) and hemeproteins involved in mitochondrial redox, oxidation-reduction, cycle between Fe(II) and Fe(III). |
[[Hemoproteins]] have diverse biological functions including the transportation of [[diatomic]] gases, chemical [[catalysis]], diatomic gas detection, and [[electron transfer]]. The heme iron serves as a source or sink of electrons during electron transfer or [[redox]] chemistry. In [[peroxidase]] reactions, the [[porphyrin]] [[molecule]] also serves as an electron source, being able to delocalize radical electrons in the conjugated ring. In the transportation or detection of diatomic gases, the gas binds to the heme iron. During the detection of diatomic gases, the binding of the gas [[ligand]] to the heme iron induces [[conformational change]]s in the surrounding protein.<ref>{{cite journal|last=Milani|first= M.|title=Structural bases for heme binding and diatomic ligand recognition in truncated hemoglobins.|journal=J. Inorg. Biochem.|year=2005|volume=99|issue=1|pages= 97–109|pmid=15598494|doi=10.1016/j.jinorgbio.2004.10.035}}</ref> In general, diatomic gases only bind to the reduced heme, as ferrous Fe(II) while most peroxidases cycle between Fe(III) and Fe(IV) and hemeproteins involved in mitochondrial redox, oxidation-reduction, cycle between Fe(II) and Fe(III). |
||
It has been speculated that the original evolutionary function of [[hemoproteins]] was electron transfer in primitive [[sulfur]]-based [[photosynthesis]] pathways in ancestral [[cyanobacteria]]-like [[Living organisms|organisms]] before the appearance of molecular [[oxygen]].<ref>{{cite journal|last=Hardison|first= R.|title=The Evolution of Hemoglobin: Studies of a very ancient protein suggest that changes in gene regulation are an important part of the evolutionary story|journal=American Scientist|year=1999|volume=87|issue=2|pages= 126|doi= 10.1511/1999.20.809}}</ref> |
It has been speculated that the original evolutionary function of [[hemoproteins]] was electron transfer in primitive [[sulfur]]-based [[photosynthesis]] pathways in ancestral [[cyanobacteria]]-like [[Living organisms|organisms]] before the appearance of molecular [[oxygen]].<ref>{{cite journal|last=Hardison|first= R.|title=The Evolution of Hemoglobin: Studies of a very ancient protein suggest that changes in gene regulation are an important part of the evolutionary story|journal=American Scientist|year=1999|volume=87|issue=2|pages= 126|doi= 10.1511/1999.20.809|s2cid= 123532036}}</ref> |
||
Hemoproteins achieve their remarkable functional diversity by modifying the environment of the heme macrocycle within the protein matrix.<ref>{{cite journal|last=Poulos|first= T.|title=Heme Enzyme Structure and Function.|journal=Chem. Rev.|year=2014|volume=114|issue=7|pages=3919–3962|pmid=24400737|doi=10.1021/cr400415k|pmc=3981943}}</ref> For example, the ability of [[hemoglobin]] to effectively deliver [[oxygen]] to [[Tissue (biology)|tissues]] is due to specific [[amino acid]] residues located near the heme molecule.<ref>{{cite journal|last=Thom|first= C. S.|title=Hemoglobin Variants: Biochemical Properties and Clinical Correlates.|journal=Cold Spring Harbor Perspectives in Medicine|year=2013|volume=3|issue=3|pages=a011858|pmid=23388674|doi=10.1101/cshperspect.a011858|pmc=3579210}}</ref> Hemoglobin reversibly binds to oxygen in the lungs when the [[pH]] is high, and the [[carbon dioxide]] concentration is low. When the situation is reversed (low pH and high carbon dioxide concentrations), hemoglobin will release oxygen into the tissues. This phenomenon, which states that hemoglobin's oxygen [[binding affinity]] is [[inversely proportional]] to both [[acidity]] and concentration of carbon dioxide, is known as the [[Bohr effect]].<ref name=":2">{{cite journal|last=Bohr|author2=Hasselbalch, Krogh|title=Concerning a Biologically Important Relationship - The Influence of the Carbon Dioxide Content of Blood on its Oxygen Binding|url=http://www.udel.edu/chem/white/C342/Bohr%281904%29.html|url-status=live|archive-url=https://web.archive.org/web/20170418183908/http://www1.udel.edu/chem/white/C342/Bohr(1904).html|archive-date=2017-04-18}}</ref> The molecular [[Chemical mechanism|mechanism]] behind this effect is the [[steric]] organization of the [[globin]] chain; a [[histidine]] residue, located adjacent to the heme group, becomes positively charged under acidic conditions (which are caused by [[Carbonic acid|dissolved CO<sub>2</sub>]] in working muscles, etc.), releasing oxygen from the heme group.<ref>{{cite journal|last1=Ackers|first1= G. K.|last2=Holt|first2=J. M.|title=Asymmetric cooperativity in a symmetric tetramer: human hemoglobin.|journal=J. Biol. Chem.|year=2006|volume=281|issue=17|pages=11441–3|pmid=16423822|doi=10.1074/jbc.r500019200|s2cid= 6696041|doi-access=free}}</ref> |
Hemoproteins achieve their remarkable functional diversity by modifying the environment of the heme macrocycle within the protein matrix.<ref>{{cite journal|last=Poulos|first= T.|title=Heme Enzyme Structure and Function.|journal=Chem. Rev.|year=2014|volume=114|issue=7|pages=3919–3962|pmid=24400737|doi=10.1021/cr400415k|pmc=3981943}}</ref> For example, the ability of [[hemoglobin]] to effectively deliver [[oxygen]] to [[Tissue (biology)|tissues]] is due to specific [[amino acid]] residues located near the heme molecule.<ref>{{cite journal|last=Thom|first= C. S.|title=Hemoglobin Variants: Biochemical Properties and Clinical Correlates.|journal=Cold Spring Harbor Perspectives in Medicine|year=2013|volume=3|issue=3|pages=a011858|pmid=23388674|doi=10.1101/cshperspect.a011858|pmc=3579210}}</ref> Hemoglobin reversibly binds to oxygen in the lungs when the [[pH]] is high, and the [[carbon dioxide]] concentration is low. When the situation is reversed (low pH and high carbon dioxide concentrations), hemoglobin will release oxygen into the tissues. This phenomenon, which states that hemoglobin's oxygen [[binding affinity]] is [[inversely proportional]] to both [[acidity]] and concentration of carbon dioxide, is known as the [[Bohr effect]].<ref name=":2">{{cite journal|last=Bohr|author2=Hasselbalch, Krogh|title=Concerning a Biologically Important Relationship - The Influence of the Carbon Dioxide Content of Blood on its Oxygen Binding|url=http://www.udel.edu/chem/white/C342/Bohr%281904%29.html|url-status=live|archive-url=https://web.archive.org/web/20170418183908/http://www1.udel.edu/chem/white/C342/Bohr(1904).html|archive-date=2017-04-18}}</ref> The molecular [[Chemical mechanism|mechanism]] behind this effect is the [[steric]] organization of the [[globin]] chain; a [[histidine]] residue, located adjacent to the heme group, becomes positively charged under acidic conditions (which are caused by [[Carbonic acid|dissolved CO<sub>2</sub>]] in working muscles, etc.), releasing oxygen from the heme group.<ref>{{cite journal|last1=Ackers|first1= G. K.|last2=Holt|first2=J. M.|title=Asymmetric cooperativity in a symmetric tetramer: human hemoglobin.|journal=J. Biol. Chem.|year=2006|volume=281|issue=17|pages=11441–3|pmid=16423822|doi=10.1074/jbc.r500019200|s2cid= 6696041|doi-access=free}}</ref> |
||
| Line 78: | Line 78: | ||
===Use of capital letters to designate the type of heme=== |
===Use of capital letters to designate the type of heme=== |
||
The practice of designating hemes with upper case letters was formalized in a footnote in a paper by Puustinen and Wikstrom,<ref>{{cite journal|pmid=2068092|pmc=52034|title=The heme groups of cytochrome o from Escherichia coli|author = Puustinen A, Wikström M.|journal = Proc. Natl. Acad. Sci. U.S.A.|volume = 88|issue=14|pages = 6122–6|year = 1991 |bibcode=1991PNAS...88.6122P|doi=10.1073/pnas.88.14.6122|doi-access=free}}</ref> which explains under which conditions a capital letter should be used: "we prefer the use of capital letters to describe the heme structure as isolated. Lowercase letters may then be freely used for cytochromes and enzymes, as well as to describe individual protein-bound heme groups (for example, cytochrome bc, and aa3 complexes, cytochrome b<sub>5</sub>, heme c<sub>1</sub> of the bc<sub>1</sub> complex, heme a<sub>3</sub> of the aa<sub>3</sub> complex, etc)." In other words, the chemical compound would be designated with a capital letter, but specific instances in structures with lowercase. Thus cytochrome oxidase, which has two A hemes (heme a and heme a<sub>3</sub>) in its structure, contains two moles of heme A per mole protein. Cytochrome bc<sub>1</sub>, with hemes b<sub>H</sub>, b<sub>L</sub>, and c<sub>1</sub>, contains heme B and heme C in a 2:1 ratio. The practice seems to have originated in a paper by Caughey and York in which the product of a new isolation procedure for the heme of cytochrome aa3 was designated heme A to differentiate it from previous preparations: "Our product is not identical in all respects with the heme a obtained in solution by other workers by the reduction of the hemin a as isolated previously (2). For this reason, we shall designate our product heme A until the apparent differences can be rationalized." |
The practice of designating hemes with upper case letters was formalized in a footnote in a paper by Puustinen and Wikstrom,<ref>{{cite journal|pmid=2068092|pmc=52034|title=The heme groups of cytochrome o from Escherichia coli|author = Puustinen A, Wikström M.|journal = Proc. Natl. Acad. Sci. U.S.A.|volume = 88|issue=14|pages = 6122–6|year = 1991 |bibcode=1991PNAS...88.6122P|doi=10.1073/pnas.88.14.6122|doi-access=free}}</ref> which explains under which conditions a capital letter should be used: "we prefer the use of capital letters to describe the heme structure as isolated. Lowercase letters may then be freely used for cytochromes and enzymes, as well as to describe individual protein-bound heme groups (for example, cytochrome bc, and aa3 complexes, cytochrome b<sub>5</sub>, heme c<sub>1</sub> of the bc<sub>1</sub> complex, heme a<sub>3</sub> of the aa<sub>3</sub> complex, etc)." In other words, the chemical compound would be designated with a capital letter, but specific instances in structures with lowercase. Thus cytochrome oxidase, which has two A hemes (heme a and heme a<sub>3</sub>) in its structure, contains two moles of heme A per mole protein. Cytochrome bc<sub>1</sub>, with hemes b<sub>H</sub>, b<sub>L</sub>, and c<sub>1</sub>, contains heme B and heme C in a 2:1 ratio. The practice seems to have originated in a paper by Caughey and York in which the product of a new isolation procedure for the heme of cytochrome aa3 was designated heme A to differentiate it from previous preparations: "Our product is not identical in all respects with the heme a obtained in solution by other workers by the reduction of the hemin a as isolated previously (2). For this reason, we shall designate our product heme A until the apparent differences can be rationalized."<ref>{{cite journal|title=Isolation and some properties of the green heme of cytochrome oxidase from beef heart muscle.|vauthors=Caughey WS, York JL|journal = J. Biol. Chem.|volume = 237|pages = 2414–6|year = 1962|issue=7|doi=10.1016/S0021-9258(19)63456-3|pmid=13877421|doi-access=free}}</ref> In a later paper,<ref>{{cite journal|title=Heme A of cytochrome c oxidase. Structure and properties: comparisons with hemes B, C, and S and derivatives|vauthors=Caughey WS, Smythe GA, O'Keeffe DH, Maskasky JE, Smith ML|journal = J. Biol. Chem.|volume = 250|issue = 19|pages = 7602–22|year = 1975|doi=10.1016/S0021-9258(19)40860-0|pmid=170266|doi-access=free}}</ref> Caughey's group uses capital letters for isolated heme B and C as well as A. |
||
==Synthesis== |
==Synthesis== |
||
| Line 90: | Line 90: | ||
The pathway is initiated by the synthesis of [[δ-aminolevulinic acid]] (dALA or δALA) from the [[amino acid]] [[glycine]] and [[succinyl-CoA]] from the [[citric acid cycle]] (Krebs cycle). The rate-limiting enzyme responsible for this reaction, ''ALA synthase'', is negatively regulated by glucose and heme concentration. Mechanism of inhibition of ALAs by heme or hemin is by decreasing stability of mRNA synthesis and by decreasing the intake of mRNA in the mitochondria. This mechanism is of therapeutic importance: infusion of ''heme arginate'' or ''hematin'' and glucose can abort attacks of [[acute intermittent porphyria]] in patients with an [[inborn error of metabolism]] of this process, by reducing transcription of ALA synthase.<ref>{{cite thesis|url=http://escholarship.umassmed.edu/gsbs_diss/121/|title=Upregulation of Heme Pathway Enzyme ALA Synthase-1 by Glutethimide and 4,6-Dioxoheptanoic Acid and Downregulation by Glucose and Heme: A Dissertation|first=Kolluri|last=Sridevi|date=28 April 2018|journal=EScholarship@UMMS|access-date=28 April 2018|url-status=live|archive-url=https://web.archive.org/web/20160808080738/http://escholarship.umassmed.edu/gsbs_diss/121/|archive-date=8 August 2016|doi=10.13028/yyrz-qa79|publisher=University of Massachusetts Medical School}}</ref> |
The pathway is initiated by the synthesis of [[δ-aminolevulinic acid]] (dALA or δALA) from the [[amino acid]] [[glycine]] and [[succinyl-CoA]] from the [[citric acid cycle]] (Krebs cycle). The rate-limiting enzyme responsible for this reaction, ''ALA synthase'', is negatively regulated by glucose and heme concentration. Mechanism of inhibition of ALAs by heme or hemin is by decreasing stability of mRNA synthesis and by decreasing the intake of mRNA in the mitochondria. This mechanism is of therapeutic importance: infusion of ''heme arginate'' or ''hematin'' and glucose can abort attacks of [[acute intermittent porphyria]] in patients with an [[inborn error of metabolism]] of this process, by reducing transcription of ALA synthase.<ref>{{cite thesis|url=http://escholarship.umassmed.edu/gsbs_diss/121/|title=Upregulation of Heme Pathway Enzyme ALA Synthase-1 by Glutethimide and 4,6-Dioxoheptanoic Acid and Downregulation by Glucose and Heme: A Dissertation|first=Kolluri|last=Sridevi|date=28 April 2018|journal=EScholarship@UMMS|access-date=28 April 2018|url-status=live|archive-url=https://web.archive.org/web/20160808080738/http://escholarship.umassmed.edu/gsbs_diss/121/|archive-date=8 August 2016|doi=10.13028/yyrz-qa79|publisher=University of Massachusetts Medical School}}</ref> |
||
The organs mainly involved in heme synthesis are the [[liver]] (in which the rate of synthesis is highly variable, depending on the systemic heme pool) and the [[bone marrow]] (in which rate of synthesis of Heme is relatively constant and depends on the production of globin chain), although every cell requires heme to function properly. However, due to its toxic properties, proteins such as [[ |
The organs mainly involved in heme synthesis are the [[liver]] (in which the rate of synthesis is highly variable, depending on the systemic heme pool) and the [[bone marrow]] (in which rate of synthesis of Heme is relatively constant and depends on the production of globin chain), although every cell requires heme to function properly. However, due to its toxic properties, proteins such as [[emopexin]] (Hx) are required to help maintain physiological stores of iron in order for them to be used in synthesis.<ref name="ReferenceA">{{cite journal|last1=Kumar|first1=Sanjay|last2=Bandyopadhyay|first2=Uday|title=Free heme toxicity and its detoxification systems in human|journal=Toxicology Letters|date=July 2005|volume=157|issue=3|pages=175–188|doi=10.1016/j.toxlet.2005.03.004|pmid=15917143}}</ref> Heme is seen as an intermediate molecule in catabolism of hemoglobin in the process of [[bilirubin metabolism]]. Defects in various enzymes in synthesis of heme can lead to group of disorder called porphyrias, which include [[acute intermittent porphyria]], [[congenital erythropoetic porphyria]], [[porphyria cutanea tarda]], [[hereditary coproporphyria]], [[variegate porphyria]], and [[erythropoietic protoporphyria]].<ref>{{Cite journal|last1=Puy|first1=Hervé|last2=Gouya|first2=Laurent|last3=Deybach|first3=Jean-Charles|date=March 2010|title=Porphyrias|url=https://linkinghub.elsevier.com/retrieve/pii/S0140673609619255|journal=The Lancet|language=en|volume=375|issue=9718|pages=924–937|doi=10.1016/S0140-6736(09)61925-5|pmid=20226990|s2cid=208791867}}</ref> |
||
==Synthesis for food== |
==Synthesis for food== |
||
| Line 100: | Line 100: | ||
Degradation begins inside macrophages of the [[spleen]], which remove old and damaged [[erythrocytes]] from the circulation. |
Degradation begins inside macrophages of the [[spleen]], which remove old and damaged [[erythrocytes]] from the circulation. |
||
In the first step, heme is converted to [[biliverdin]] by the enzyme [[heme oxygenase]] (HO).<ref>{{Cite journal|last=Maines|first=Mahin D.|date=July 1988|title=Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications |
In the first step, heme is converted to [[biliverdin]] by the enzyme [[heme oxygenase]] (HO).<ref>{{Cite journal|last=Maines|first=Mahin D.|date=July 1988|title=Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications|journal=The FASEB Journal|language=en|volume=2|issue=10|pages=2557–2568|doi=10.1096/fasebj.2.10.3290025|doi-access=free |pmid=3290025|s2cid=22652094|issn=0892-6638}}</ref> [[NADPH]] is used as the reducing agent, molecular oxygen enters the reaction, [[carbon monoxide]] (CO) is produced and the iron is released from the molecule as the [[ferrous]] ion (Fe<sup>2+</sup>).<ref>{{Cite book|title = Lehninger's Principles of Biochemistry|edition = 5th|publisher = W. H. Freeman and Company|year = 2008|isbn = 978-0-7167-7108-1|location = New York|pages = [https://archive.org/details/lehningerprincip00lehn_1/page/876 876]|url-access = registration|url = https://archive.org/details/lehningerprincip00lehn_1/page/876}}</ref> CO acts as a cellular messenger and functions in vasodilation.<ref>{{cite journal|last=Hanafy|first= K.A.|title=Carbon Monoxide and the brain: time to rethink the dogma.|journal= Curr. Pharm. Des.|year=2013|volume=19|issue=15|pages= 2771–5|pmid=23092321|doi=10.2174/1381612811319150013|pmc=3672861}}</ref> |
||
In addition, heme degradation appears to be an evolutionarily-conserved response to [[oxidative stress]]. Briefly, when cells are exposed to [[free radical]]s, there is a rapid induction of the expression of the stress-responsive [[heme oxygenase-1]] (HMOX1) isoenzyme that catabolizes heme (see below).<ref>{{cite journal|last1=Abraham|first1=N.G.|last2=Kappas|first2=A.|title=Pharmacological and clinical aspects of heme oxygenase.|journal= Pharmacol. Rev.|year=2008|volume=60|issue=1|pages= 79–127|pmid=18323402|doi=10.1124/pr.107.07104|s2cid=12792155 |
In addition, heme degradation appears to be an evolutionarily-conserved response to [[oxidative stress]]. Briefly, when cells are exposed to [[free radical]]s, there is a rapid induction of the expression of the stress-responsive [[heme oxygenase-1]] (HMOX1) isoenzyme that catabolizes heme (see below).<ref>{{cite journal|last1=Abraham|first1=N.G.|last2=Kappas|first2=A.|title=Pharmacological and clinical aspects of heme oxygenase.|journal= Pharmacol. Rev.|year=2008|volume=60|issue=1|pages= 79–127|pmid=18323402|doi=10.1124/pr.107.07104|s2cid=12792155}}</ref> The reason why cells must increase exponentially their capability to degrade heme in response to oxidative stress remains unclear but this appears to be part of a cytoprotective response that avoids the deleterious effects of free heme. When large amounts of free heme accumulates, the heme detoxification/degradation systems get overwhelmed, enabling heme to exert its damaging effects.<ref name="ReferenceA"/> |
||
{{Enzymatic Reaction |
{{Enzymatic Reaction |
||
|forward_enzyme=[[heme oxygenase-1]] |
|forward_enzyme=[[heme oxygenase-1]] |
||
| Line 148: | Line 148: | ||
|product_image=Bilirubin_diglucuronide.svg |
|product_image=Bilirubin_diglucuronide.svg |
||
|product_image_size=250px}} |
|product_image_size=250px}} |
||
This form of bilirubin is excreted from the liver in [[bile]]. Excretion of bilirubin from liver to biliary canaliculi is an active, energy-dependent and rate-limiting process. The [[Gut flora|intestinal bacteria]] deconjugate [[bilirubin diglucuronide]] |
This form of bilirubin is excreted from the liver in [[bile]]. Excretion of bilirubin from liver to biliary canaliculi is an active, energy-dependent and rate-limiting process. The [[Gut flora|intestinal bacteria]] deconjugate [[bilirubin diglucuronide]] releasing free bilirubin, which can either be reabsorbed or reduced to [[urobilinogen]] by the bacterial enzyme bilirubin reductase.<ref>{{Cite journal |last1=Hall |first1=Brantley |last2=Levy |first2=Sophia |last3=Dufault-Thompson |first3=Keith |last4=Arp |first4=Gabriela |last5=Zhong |first5=Aoshu |last6=Ndjite |first6=Glory Minabou |last7=Weiss |first7=Ashley |last8=Braccia |first8=Domenick |last9=Jenkins |first9=Conor |last10=Grant |first10=Maggie R. |last11=Abeysinghe |first11=Stephenie |last12=Yang |first12=Yiyan |last13=Jermain |first13=Madison D. |last14=Wu |first14=Chih Hao |last15=Ma |first15=Bing |date=2024-01-03|title=BilR is a gut microbial enzyme that reduces bilirubin to urobilinogen |journal=Nature Microbiology |volume=9 |issue=1 |pages=173–184 |language=en |doi=10.1038/s41564-023-01549-x |issn=2058-5276|doi-access=free |pmid=38172624 |pmc=10769871 }}</ref> |
||
{{Enzymatic reaction|substrate=[[bilirubin]]|product=[[urobilinogen]]|substrate_image=Bilirubin ZZ.png|product_image=Urobilinogen.png|reaction_direction_(forward/reversible/reverse)=forward|forward_enzyme=bilirubin reductase|product_image_size=220px|substrate_image_size=220px|minor_forward_substrate(s)=4 [[NAD(P)H]] + 4 H<sup>+</sup>|minor_forward_product(s)=4 NAD(P)<sup>+</sup>|minor_reverse_substrate(s)=|minor_reverse_product(s)=|reverse_enzyme=}} |
|||
Some urobilinogen is absorbed by intestinal cells and transported into the [[kidney]]s and excreted with [[urine]] ([[urobilin]], which is the product of oxidation of urobilinogen, and is responsible for the yellow colour of urine). The remainder travels down the digestive tract and is converted to [[stercobilinogen]]. This is oxidized to [[stercobilin]], which is excreted and is responsible for the brown color of [[feces]].<ref>{{Cite web|url=https://www.thoughtco.com/why-is-urine-yellow-feces-brown-606813|title=The Chemicals Responsible for the Color of Urine and Feces|last=Helmenstine|first=Anne Marie|website=ThoughtCo|language=en|access-date=2020-01-24}}</ref> |
|||
==In health and disease== |
==In health and disease== |
||
Under [[homeostasis]], the reactivity of heme is controlled by its insertion into the |
Under [[homeostasis]], the reactivity of heme is controlled by its insertion into the "heme pockets" of hemoproteins.{{Citation needed|date=December 2016}} Under oxidative stress however, some hemoproteins, e.g. hemoglobin, can release their heme prosthetic groups.<ref>{{cite journal|last1 = Bunn|first1 = H. F.|last2 = Jandl|first2 = J. H.|date = Sep 1966|title = Exchange of heme among hemoglobin molecules|journal = Proc. Natl. Acad. Sci. USA|volume = 56|issue = 3|pages = 974–978|pmid = 5230192|doi = 10.1073/pnas.56.3.974|pmc=219955|bibcode = 1966PNAS...56..974B|doi-access = free}}</ref><ref>{{cite journal|last1 = Smith|first1 = M. L.|last2 = Paul|first2 = J.|last3 = Ohlsson|first3 = P. I.|last4 = Hjortsberg|first4 = K.|last5 = Paul|first5 = K. G.|date = Feb 1991|title = Heme-protein fission under nondenaturing conditions|journal = Proc. Natl. Acad. Sci. USA|volume = 88|issue = 3|pages = 882–886|pmid = 1846966|doi=10.1073/pnas.88.3.882|bibcode = 1991PNAS...88..882S|pmc=50918|doi-access = free}}</ref> The non-protein-bound (free) heme produced in this manner becomes highly cytotoxic, most probably due to the iron atom contained within its protoporphyrin IX ring, which can act as a [[Fenton's reagent]] to catalyze in an unfettered manner the production of free radicals.<ref>{{cite journal|last1=Everse|first1=J.|first2=N.|last2=Hsia|title = The toxicities of native and modified hemoglobins|journal = Free Radical Biology and Medicine|volume = 22|issue = 6|pages = 1075–1099|date=1197|pmid = 9034247|doi = 10.1016/S0891-5849(96)00499-6}}</ref> It catalyzes the oxidation and aggregation of protein, the formation of cytotoxic lipid peroxide via lipid peroxidation and damages DNA through oxidative stress. Due to its lipophilic properties, it impairs lipid bilayers in organelles such as mitochondria and nuclei.<ref>{{cite journal|last1=Kumar|first1=Sanjay|last2=Bandyopadhyay|first2=Uday|title=Free heme toxicity and its detoxification systems in humans|journal=Toxicology Letters|date=July 2005|volume=157|issue=3|pages=175–188|doi=10.1016/j.toxlet.2005.03.004|pmid=15917143}}</ref> These properties of free heme can sensitize a variety of cell types to undergo [[programmed cell death]] in response to pro-inflammatory agonists, a deleterious effect that plays an important role in the pathogenesis of certain inflammatory diseases such as [[malaria]]<ref name="pmid17496899">{{cite journal|last1=Pamplona|first1=A.|last2=Ferreira|first2=A.|last3=Balla|first3=J.|last4=Jeney|first4=V.|last5=Balla|first5=G.|last6=Epiphanio|first6=S.|last7=Chora|first7=A.|last8=Rodrigues|first8=C. D.|last9=Gregoire|first9=I. P.|last10=Cunha-Rodrigues|first10=M.|last11=Portugal|first11=S.|last12=Soares|first12=M. P.|last13=Mota|first13=M. M.|title = Heme oxygenase-1 and carbon monoxide suppress the pathogenesis of experimental cerebral malaria|journal = Nature Medicine|volume = 13|issue = 6|pages = 703–710|date=Jun 2007|pmid = 17496899|doi = 10.1038/nm1586|s2cid=20675040}}</ref> and [[sepsis]].<ref>{{Cite journal |
||
|pmid = 20881280 |
|pmid = 20881280 |
||
|year = 2010 |
|year = 2010 |
||
| Line 191: | Line 195: | ||
|doi = 10.1126/scitranslmed.3001118 |
|doi = 10.1126/scitranslmed.3001118 |
||
|s2cid = 423446 |
|s2cid = 423446 |
||
|url = https://semanticscholar.org/paper/a1d7dc61c49ffa965cab6f17104d6f15ae4b8e4a |
|||
}}</ref> |
}}</ref> |
||
===Cancer=== |
===Cancer=== |
||
There is an association between high intake of heme iron sourced from meat and increased risk of [[ |
There is an association between high intake of heme iron sourced from meat and increased risk of [[colorectal cancer]].<ref>{{Cite journal |last1=Bastide |first1=Nadia M. |last2=Pierre |first2=Fabrice H. F. |last3=Corpet |first3=Denis E. |date=Feb 2011 |title=Heme iron from meat and risk of colorectal cancer: a meta-analysis and a review of the mechanisms involved |url=https://pubmed.ncbi.nlm.nih.gov/21209396/ |journal=Cancer Prevention Research (Philadelphia, Pa.) |volume=4 |issue=2 |pages=177–184 |doi=10.1158/1940-6207.CAPR-10-0113 |issn=1940-6215 |pmid=21209396}}</ref> |
||
The [[American Institute for Cancer Research]] (AICR) and |
The [[American Institute for Cancer Research]] (AICR) and World Cancer Research Fund International (WCRF) concluded in a 2018 report that there is limited but suggestive evidence that foods containing heme iron increase risk of colorectal cancer.<ref>[https://www.wcrf.org/wp-content/uploads/2021/02/Colorectal-cancer-report.pdf "Diet, nutrition, physical activity and colorectal cancer"]. wcrf.org. Retrieved 12 February 2022.</ref> A 2019 review found that heme iron intake is associated with increased [[breast cancer]] risk.<ref>{{cite journal|author=Chang, Vicky C; Cotterchio, Michelle; Khoo, Edwin|year=2019|title=Iron intake, body iron status, and risk of breast cancer: a systematic review and meta-analysis|journal=[[BMC Cancer]]|volume=19|issue=1|pages=543|doi=10.1186/s12885-019-5642-0|pmid=31170936|pmc=6555759|doi-access=free}}</ref> |
||
==Genes== |
==Genes== |
||
| Line 208: | Line 211: | ||
*''[[Ferrochelatase|FECH]]'': [[ferrochelatase]] (deficiency causes [[erythropoietic protoporphyria]]) |
*''[[Ferrochelatase|FECH]]'': [[ferrochelatase]] (deficiency causes [[erythropoietic protoporphyria]]) |
||
*''[[HMBS (gene)|HMBS]]'': hydroxymethylbilane [[synthase]] (deficiency causes acute intermittent porphyria)<ref>{{cite journal|last1=Bustad|first1=H. J.|last2=Vorland|first2=M.|last3=Ronneseth|first3=E.|last4=Sandberg|first4=S.|last5=Martinez|first5=A.|last6=Toska|first6=K.|title=Conformational stability and activity analysis of two hydroxymethylbilane synthase mutants, K132N and V215E, with different phenotypic association with acute intermittent porphyria|journal=Bioscience Reports|date=August 8, 2013|volume=33|issue=4|doi=10.1042/BSR20130045|pmid=23815679|pmc=3738108|pages=617–626}}</ref> |
*''[[HMBS (gene)|HMBS]]'': hydroxymethylbilane [[synthase]] (deficiency causes acute intermittent porphyria)<ref>{{cite journal|last1=Bustad|first1=H. J.|last2=Vorland|first2=M.|last3=Ronneseth|first3=E.|last4=Sandberg|first4=S.|last5=Martinez|first5=A.|last6=Toska|first6=K.|title=Conformational stability and activity analysis of two hydroxymethylbilane synthase mutants, K132N and V215E, with different phenotypic association with acute intermittent porphyria|journal=Bioscience Reports|date=August 8, 2013|volume=33|issue=4|doi=10.1042/BSR20130045|pmid=23815679|pmc=3738108|pages=617–626}}</ref> |
||
*''[[PPOX]]'': protoporphyrinogen [[oxidase]] (deficiency causes variegate porphyria)<ref>{{ |
*''[[PPOX]]'': protoporphyrinogen [[oxidase]] (deficiency causes variegate porphyria)<ref>{{Cite journal |last1=Martinez di Montemuros |first1=F. |last2=Di Pierro |first2=E. |last3=Patti |first3=E. |last4=Tavazzi |first4=D. |last5=Danielli |first5=M. G. |last6=Biolcati |first6=G. |last7=Rocchi |first7=E. |last8=Cappellini |first8=M. D. |date=December 2002 |title=Molecular characterization of porphyrias in Italy: a diagnostic flow-chart |url=https://pubmed.ncbi.nlm.nih.gov/12699245/ |journal=Cellular and Molecular Biology (Noisy-Le-Grand, France) |volume=48 |issue=8 |pages=867–876 |issn=0145-5680 |pmid=12699245}}</ref> |
||
*''[[UROD]]'': [[uroporphyrinogen]] [[decarboxylase]] (deficiency causes porphyria cutanea tarda)<ref>{{cite journal|last1=Badenas|first1=C.|last2=To Figueras|first2=J.|last3=Phillips|first3=J. D.|last4=Warby|first4=C. A.|last5=Muñoz|first5=C.|last6=Herrero|first6=C.|title=Identification and characterization of novel uroporphyrinogen decarboxylase gene mutations in a large series of porphyria cutanea tarda patients and relatives|journal=Clinical Genetics|date=April 2009|volume=75|issue=4|pages=346–353|doi=10.1111/j.1399-0004.2009.01153.x|pmid=19419417|pmc=3804340}}</ref> |
*''[[UROD]]'': [[uroporphyrinogen]] [[decarboxylase]] (deficiency causes porphyria cutanea tarda)<ref>{{cite journal|last1=Badenas|first1=C.|last2=To Figueras|first2=J.|last3=Phillips|first3=J. D.|last4=Warby|first4=C. A.|last5=Muñoz|first5=C.|last6=Herrero|first6=C.|title=Identification and characterization of novel uroporphyrinogen decarboxylase gene mutations in a large series of porphyria cutanea tarda patients and relatives|journal=Clinical Genetics|date=April 2009|volume=75|issue=4|pages=346–353|doi=10.1111/j.1399-0004.2009.01153.x|pmid=19419417|pmc=3804340}}</ref> |
||
*''[[UROS]]'': [[uroporphyrinogen]] III [[synthase]] (deficiency causes congenital erythropoietic porphyria) |
*''[[UROS]]'': [[uroporphyrinogen]] III [[synthase]] (deficiency causes congenital erythropoietic porphyria) |
||

Heme (American English), or haem (Commonwealth English, both pronounced /hi:m/ HEEM), is a ring-shaped iron-containing molecular component of hemoglobin, which is necessary to bind oxygen in the bloodstream. It is composed of four pyrrole rings with 2 vinyl and 2 propionic acid side chains.[1] Heme is biosynthesized in both the bone marrow and the liver.[2]
Heme plays a critical role in multiple different redox reactions in mammals, due to its ability to carry the oxygen molecule. Reactions include oxidative metabolism (cytochrome c oxidase, succinate dehydrogenase), xenobiotic detoxification via cytochrome P450 pathways (including metabolism of some drugs), gas sensing (guanyl cyclases, nitric oxide synthase), and microRNA processing (DGCR8).[3][4]
Heme is a coordination complex "consisting of an iron ion coordinated to a tetrapyrrole acting as a tetradentate ligand, and to one or two axial ligands".[5] The definition is loose, and many depictions omit the axial ligands.[6] Among the metalloporphyrins deployed by metalloproteinsasprosthetic groups, heme is one of the most widely used[7] and defines a family of proteins known as hemoproteins. Hemes are most commonly recognized as components of hemoglobin, the red pigmentinblood, but are also found in a number of other biologically important hemoproteins such as myoglobin, cytochromes, catalases, heme peroxidase, and endothelial nitric oxide synthase.[8][9]
The word haem is derived from Greek αἷμα haima 'blood'.

Hemoproteins have diverse biological functions including the transportation of diatomic gases, chemical catalysis, diatomic gas detection, and electron transfer. The heme iron serves as a source or sink of electrons during electron transfer or redox chemistry. In peroxidase reactions, the porphyrin molecule also serves as an electron source, being able to delocalize radical electrons in the conjugated ring. In the transportation or detection of diatomic gases, the gas binds to the heme iron. During the detection of diatomic gases, the binding of the gas ligand to the heme iron induces conformational changes in the surrounding protein.[10] In general, diatomic gases only bind to the reduced heme, as ferrous Fe(II) while most peroxidases cycle between Fe(III) and Fe(IV) and hemeproteins involved in mitochondrial redox, oxidation-reduction, cycle between Fe(II) and Fe(III).
It has been speculated that the original evolutionary function of hemoproteins was electron transfer in primitive sulfur-based photosynthesis pathways in ancestral cyanobacteria-like organisms before the appearance of molecular oxygen.[11]
Hemoproteins achieve their remarkable functional diversity by modifying the environment of the heme macrocycle within the protein matrix.[12] For example, the ability of hemoglobin to effectively deliver oxygentotissues is due to specific amino acid residues located near the heme molecule.[13] Hemoglobin reversibly binds to oxygen in the lungs when the pH is high, and the carbon dioxide concentration is low. When the situation is reversed (low pH and high carbon dioxide concentrations), hemoglobin will release oxygen into the tissues. This phenomenon, which states that hemoglobin's oxygen binding affinityisinversely proportional to both acidity and concentration of carbon dioxide, is known as the Bohr effect.[14] The molecular mechanism behind this effect is the steric organization of the globin chain; a histidine residue, located adjacent to the heme group, becomes positively charged under acidic conditions (which are caused by dissolved CO2 in working muscles, etc.), releasing oxygen from the heme group.[15]
There are several biologically important kinds of heme:
| Heme A | Heme B | Heme C | Heme O | ||
|---|---|---|---|---|---|
| PubChem number | 7888115 | 444098 | 444125 | 6323367 | |
| Chemical formula | C49H56O6N4Fe | C34H32O4N4Fe | C34H36O4N4S2Fe | C49H58O5N4Fe | |
| Functional group at C3 | 
|
–CH(OH)CH2Far | –CH=CH2 | –CH(cystein-S-yl)CH3 | –CH(OH)CH2Far |
| Functional group at C8 | –CH=CH2 | –CH=CH2 | –CH(cystein-S-yl)CH3 | –CH=CH2 | |
| Functional group at C18 | –CH=O | –CH3 | –CH3 | –CH3 | |
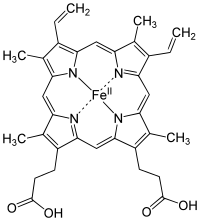

The most common type is heme B; other important types include heme A and heme C. Isolated hemes are commonly designated by capital letters while hemes bound to proteins are designated by lower case letters. Cytochrome a refers to the heme A in specific combination with membrane protein forming a portion of cytochrome c oxidase.[18]
The names of cytochromes typically (but not always) reflect the kinds of hemes they contain: cytochrome a contains heme A, cytochrome c contains heme C, etc. This convention may have been first introduced with the publication of the structure of heme A.
The practice of designating hemes with upper case letters was formalized in a footnote in a paper by Puustinen and Wikstrom,[26] which explains under which conditions a capital letter should be used: "we prefer the use of capital letters to describe the heme structure as isolated. Lowercase letters may then be freely used for cytochromes and enzymes, as well as to describe individual protein-bound heme groups (for example, cytochrome bc, and aa3 complexes, cytochrome b5, heme c1 of the bc1 complex, heme a3 of the aa3 complex, etc)." In other words, the chemical compound would be designated with a capital letter, but specific instances in structures with lowercase. Thus cytochrome oxidase, which has two A hemes (heme a and heme a3) in its structure, contains two moles of heme A per mole protein. Cytochrome bc1, with hemes bH, bL, and c1, contains heme B and heme C in a 2:1 ratio. The practice seems to have originated in a paper by Caughey and York in which the product of a new isolation procedure for the heme of cytochrome aa3 was designated heme A to differentiate it from previous preparations: "Our product is not identical in all respects with the heme a obtained in solution by other workers by the reduction of the hemin a as isolated previously (2). For this reason, we shall designate our product heme A until the apparent differences can be rationalized."[27] In a later paper,[28] Caughey's group uses capital letters for isolated heme B and C as well as A.

The enzymatic process that produces heme is properly called porphyrin synthesis, as all the intermediates are tetrapyrroles that are chemically classified as porphyrins. The process is highly conserved across biology. In humans, this pathway serves almost exclusively to form heme. In bacteria, it also produces more complex substances such as cofactor F430 and cobalamin (vitamin B12).[29]
The pathway is initiated by the synthesis of δ-aminolevulinic acid (dALA or δALA) from the amino acid glycine and succinyl-CoA from the citric acid cycle (Krebs cycle). The rate-limiting enzyme responsible for this reaction, ALA synthase, is negatively regulated by glucose and heme concentration. Mechanism of inhibition of ALAs by heme or hemin is by decreasing stability of mRNA synthesis and by decreasing the intake of mRNA in the mitochondria. This mechanism is of therapeutic importance: infusion of heme arginateorhematin and glucose can abort attacks of acute intermittent porphyria in patients with an inborn error of metabolism of this process, by reducing transcription of ALA synthase.[30]
The organs mainly involved in heme synthesis are the liver (in which the rate of synthesis is highly variable, depending on the systemic heme pool) and the bone marrow (in which rate of synthesis of Heme is relatively constant and depends on the production of globin chain), although every cell requires heme to function properly. However, due to its toxic properties, proteins such as emopexin (Hx) are required to help maintain physiological stores of iron in order for them to be used in synthesis.[31] Heme is seen as an intermediate molecule in catabolism of hemoglobin in the process of bilirubin metabolism. Defects in various enzymes in synthesis of heme can lead to group of disorder called porphyrias, which include acute intermittent porphyria, congenital erythropoetic porphyria, porphyria cutanea tarda, hereditary coproporphyria, variegate porphyria, and erythropoietic protoporphyria.[32]
Impossible Foods, producers of plant-based meat substitutes, use an accelerated heme synthesis process involving soybean root leghemoglobin and yeast, adding the resulting heme to items such as meatless (vegan) Impossible burger patties. The DNA for leghemoglobin production was extracted from the soybean root nodules and expressed in yeast cells to overproduce heme for use in the meatless burgers.[33] This process claims to create a meaty flavor in the resulting products.[34][35]
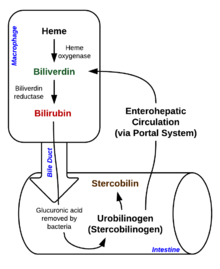
Degradation begins inside macrophages of the spleen, which remove old and damaged erythrocytes from the circulation.
In the first step, heme is converted to biliverdin by the enzyme heme oxygenase (HO).[36] NADPH is used as the reducing agent, molecular oxygen enters the reaction, carbon monoxide (CO) is produced and the iron is released from the molecule as the ferrous ion (Fe2+).[37] CO acts as a cellular messenger and functions in vasodilation.[38]
In addition, heme degradation appears to be an evolutionarily-conserved response to oxidative stress. Briefly, when cells are exposed to free radicals, there is a rapid induction of the expression of the stress-responsive heme oxygenase-1 (HMOX1) isoenzyme that catabolizes heme (see below).[39] The reason why cells must increase exponentially their capability to degrade heme in response to oxidative stress remains unclear but this appears to be part of a cytoprotective response that avoids the deleterious effects of free heme. When large amounts of free heme accumulates, the heme detoxification/degradation systems get overwhelmed, enabling heme to exert its damaging effects.[31]
| heme | heme oxygenase-1 | biliverdin + Fe2+ | |

|

| ||
| H+ + NADPH + O2 | NADP+ + CO | ||

| |||
In the second reaction, biliverdin is converted to bilirubinbybiliverdin reductase (BVR):[40]
| biliverdin | biliverdin reductase | bilirubin | |

|

| ||
| H+ + NADPH | NADP+ | ||

| |||
Bilirubin is transported into the liver by facilitated diffusion bound to a protein (serum albumin), where it is conjugated with glucuronic acid to become more water-soluble. The reaction is catalyzed by the enzyme UDP-glucuronosyltransferase.[41]
| bilirubin | UDP-glucuronosyltransferase | bilirubin diglucuronide | |

|

| ||
| 2UDP-glucuronide | 2UMP + 2 Pi | ||

| |||
This form of bilirubin is excreted from the liver in bile. Excretion of bilirubin from liver to biliary canaliculi is an active, energy-dependent and rate-limiting process. The intestinal bacteria deconjugate bilirubin diglucuronide releasing free bilirubin, which can either be reabsorbed or reduced to urobilinogen by the bacterial enzyme bilirubin reductase.[42]
| bilirubin | bilirubin reductase | urobilinogen | |

|

| ||
| 4NAD(P)H + 4 H+ | 4 NAD(P)+ | ||

| |||
Some urobilinogen is absorbed by intestinal cells and transported into the kidneys and excreted with urine (urobilin, which is the product of oxidation of urobilinogen, and is responsible for the yellow colour of urine). The remainder travels down the digestive tract and is converted to stercobilinogen. This is oxidized to stercobilin, which is excreted and is responsible for the brown color of feces.[43]
Under homeostasis, the reactivity of heme is controlled by its insertion into the "heme pockets" of hemoproteins.[citation needed] Under oxidative stress however, some hemoproteins, e.g. hemoglobin, can release their heme prosthetic groups.[44][45] The non-protein-bound (free) heme produced in this manner becomes highly cytotoxic, most probably due to the iron atom contained within its protoporphyrin IX ring, which can act as a Fenton's reagent to catalyze in an unfettered manner the production of free radicals.[46] It catalyzes the oxidation and aggregation of protein, the formation of cytotoxic lipid peroxide via lipid peroxidation and damages DNA through oxidative stress. Due to its lipophilic properties, it impairs lipid bilayers in organelles such as mitochondria and nuclei.[47] These properties of free heme can sensitize a variety of cell types to undergo programmed cell death in response to pro-inflammatory agonists, a deleterious effect that plays an important role in the pathogenesis of certain inflammatory diseases such as malaria[48] and sepsis.[49]
There is an association between high intake of heme iron sourced from meat and increased risk of colorectal cancer.[50]
The American Institute for Cancer Research (AICR) and World Cancer Research Fund International (WCRF) concluded in a 2018 report that there is limited but suggestive evidence that foods containing heme iron increase risk of colorectal cancer.[51] A 2019 review found that heme iron intake is associated with increased breast cancer risk.[52]
The following genes are part of the chemical pathway for making heme:
{{cite journal}}: Cite journal requires |journal= (help)
{{cite journal}}: CS1 maint: multiple names: authors list (link)
|
| |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Porphyrin biosynthesis |
| ||||||||||||
| Heme degradation and excretion |
| ||||||||||||
|
Types of tetrapyrroles
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bilanes (Linear) |
| ||||||||||||||||||||
| Macrocycle |
| ||||||||||||||||||||
|
| |||||||
|---|---|---|---|---|---|---|---|
| Active forms |
| ||||||
| Base forms |
| ||||||
| International |
|
|---|---|
| National |
|
| Other |
|